Running CHAMP¶
CHAMP uses the quick hull algorithm to find the intersection of the space above all of the planes representing the input set of partitions as shown in Single Layer and Multilayer. There are many great community detection tools available (both in python and other langauges) that can be used to generate the starting collection of partitions which CHAMP can be applied to. We have incorporated a python version of the louvain algorithm into CHAMP to identify partitions on the basis of modularity maximization which CHAMP can then be applied to. See Louvain Extension.
Below we detail running of CHAMP for two scenarios:
- Starting from an ensemble of partitions (without the corresponding partition coefficients for calculating the convex hull).
- Starting with the partitions and the coefficients precalculated.
Starting from Partitions¶
If the partitions were generated using a modularity based community detection method, it’s better to calculate the coefficients while optimizing the communities and feed these into CHAMP directly. This is especially true, if the community detection is being performed in parallel. However, if the partitions were generated using some other form of community detection algorithm, we provide a method to compute these coefficients directly and allow for parallelization of this process on supported machines.
-
champ.champ_functions.create_coefarray_from_partitions(partition_array, A_mat, P_mat, C_mat=None, nprocesses=0)[source]¶ Parameters: - partition_array – Each row is one of M partitions of the network with N nodes. Community labels must be hashable.
- A_mat – Interlayer (single layer) adjacency matrix
- P_mat – Matrix representing null model of connectivity (i.e configuration model - \(\frac{k_ik_j}{2m}\)
- C_mat – Optional matrix representing interlayer connectivity
- nprocesses (int) – Optional number of processes to use (0 or 1 for single core)
Returns: size \(M\times\text{Dim}\) array of coefficients for each partition. Dim can be 2 (single layer) or 3 (multilayer)
Coeffients from Partitions Example¶
import champ
import numpy as np
import matplotlib.pyplot as plt
import igraph as ig
rand_er_graph=ig.Graph.Erdos_Renyi(n=1000,p=.05)
for i in range(100):
ncoms=np.random.choice(range(1,30),size=1)[0]
if i==0:
rand_partitions=np.random.choice(range(ncoms),replace=True,size=(1,1000))
else:
rand_partitions=np.concatenate([rand_partitions,np.random.choice(range(ncoms),replace=True,size=(1,1000))])
print(rand_partitions.shape)
#get the adjacency of ER graph
A_mat=np.array(rand_er_graph.get_adjacency().data)
#create null model matrix
P_mat=np.outer(rand_er_graph.degree(),rand_er_graph.degree())
## Create the array of coefficients for the partitions
coeff_array=champ.champ_functions.create_coefarray_from_partitions(A_mat=A_mat,
P_mat=P_mat,
partition_array=rand_partitions)
#Calculate the intersection of all of the halfspaces. These are the partitions that form the CHAMP set.
ind2doms=champ.champ_functions.get_intersection(coef_array=coeff_array)
print(ind2doms)
Starting from Partition Coefficients¶
In practice, it is often easier to calculate the coefficients while running performing the community detection to generate the input ensemble of partitions, especially if these partitions are being generated in parallel. If these have been generated already, one can apply CHAMP directly via the following call. The same command is used in both the Single Layer and Multilayer context, with the output determined automatically by the number of coefficients supplied in the input array.
-
champ.champ_functions.get_intersection(coef_array, max_pt=None)[source]¶ - Calculate the intersection of the halfspaces (planes) that form the convex hull
Parameters: - coef_array (array) – NxM array of M coefficients across each row representing N partitions
- max_pt ((float,float) or float) – Upper bound for the domains (in the xy plane). This will restrict the convex hull to be within the specified range of gamma/omega (such as the range of parameters originally searched using Louvain).
Returns: dictionary mapping the index of the elements in the convex hull to the points defining the boundary of the domain
Applying CHAMP to Coefficients Array Example¶
import champ
import matplotlib.pyplot as plt
#generate random coefficent matrices
coeffs=champ.get_random_halfspaces(100,dim=3)
ind_2_dom=champ.get_intersection(coeffs)
ax=champ.plot_2d_domains(ind_2_dom)
plt.show()
Output [1] :
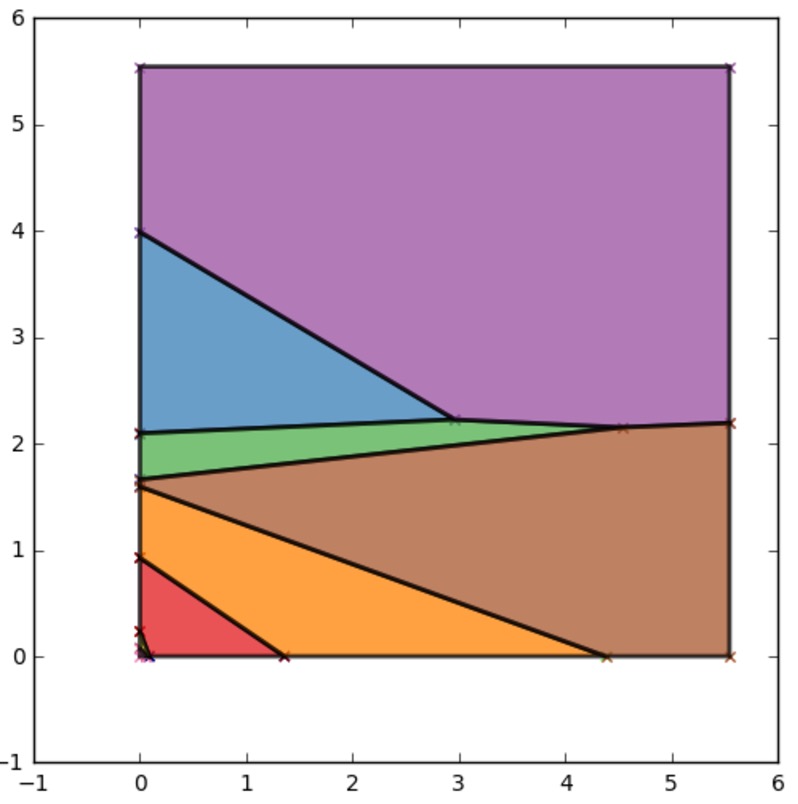
| [1] | Note that actual output might differ due to random seeding. |